Operon Definition
An operon is a cluster of functionally-related genes that are controlled by a shared operator. Operons consist of multiple genes grouped together with a promoter and an operator. Operons are present in prokaryotes (bacteria and archaea), but are absent in eukaryotes.
In some situations multiple operons are controlled by the same regulatory protein; in these cases the operons form a regulon. Operons were first identified as a mode of gene expression control in 1961 by François Jacob and Jacques Monod.
Operon Structure
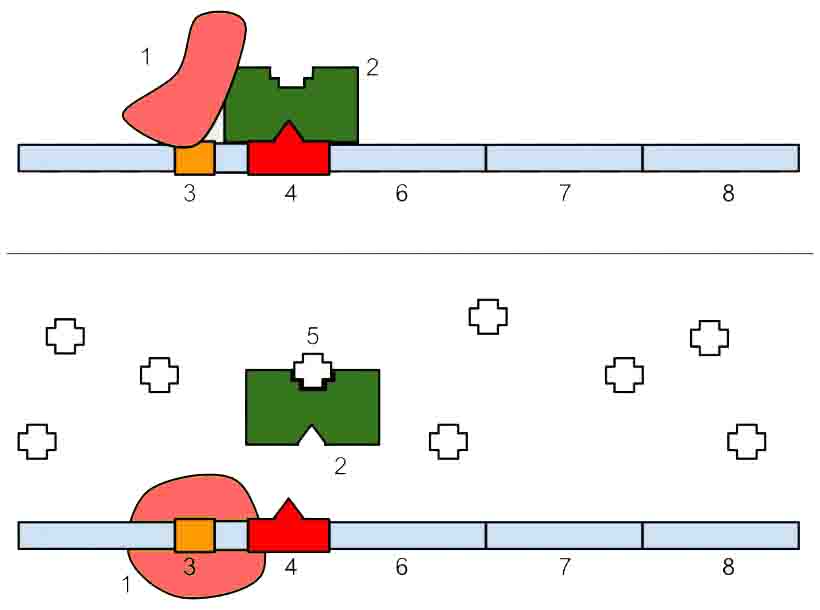
Operons are regions of DNA that contain clusters of related genes. They are made up of a promoter region, an operator, and multiple related genes. The operator can be located either within the promoter or between the promoter and the genes.
RNA polymerase initiates transcription by binding to the promoter region. The location of the operator is important as its regulation either allows or prevents transcription of the genes into mRNA.
Operon Function
An operon is a complete package for gene expression and synthesis of polypeptides. By combining the related genes, all polypeptides required for a specific function are synthesized in response to a single stimulus.
For example, the bacterium Escherichia coli contains a number of genes clustered into operons and regulons: the Lac operon which is involved in lactose degradation, the Trp operon which is involved in tryptophan biosynthesis, and the His operon which is involved in histidine biosynthesis. These operons are turned on when the gene products are needed.
Operons can be under negative or positive control. Negative control involves turning off the operon in the presence of a repressor; this can be either repressible or inducible. A repressible operon is one that is usually on but which can be repressed in the presence of a repressor molecule. The repressor binds to the operator in such a way that the movement or binding of RNA polymerase is blocked and transcription cannot proceed.
An inducible operon is one that is usually off. In the absence of an inducer the operator is blocked by a repressor molecule. When the inducer is present it interacts with the repressor protein, releasing it from the operator and allowing transcription to proceed.
Repressible operons are generally involved in anabolic pathways, or the synthesis of an essential component, while inducible operons are generally involved in catabolic pathways, or the breakdown of a nutrient. Positive control of an operon is when gene expression is stimulated by the presence of a regulatory protein.
Lac Operon
The Lac operon is the classic operon example, and is responsible for the degradation of the milk protein lactose. The Lac operon is an inducible operon; in the absence of lactose the operator is blocked by a repressor protein. The operon is made up of a promoter with operator, and three genes (lacZ, lacY, and lacA) which encode β-galactosidase, permease, and transacetylase.
The three genes are involved in the breakdown of lactose into its metabolites: β-galactosidase breaks lactose down into glucose and galactose, while the other two proteins aid in the metabolic process. The expression of the Lac operon is controlled by the regulatory gene lacI, located immediately adjacent to the promoter region. LacI encodes an allosteric repressor protein that keep the Lac operon “off”.
In order for the Lac operon to be turned on, an inducer molecule must inactivate the repressor protein. The inducer molecule in this system is allolactose, an isomer of lactose. When lactose and its isomer are present in the cell, allolactose will bind to allosteric sites on the repressor protein, changing its conformation and rendering it inactive.
As the repressor protein detaches from the operator, RNA polymerase can bind to the promoter, transcription can occur, and the three lactose degradation genes can be synthesized.
The Lac operon is also under positive gene regulation. While the removal of the repressor protein in the presence of lactose is required for synthesis of the lacZ, lacY, and lacA genes, the gene expression will remain low. The level of gene expression is controlled by the amount of the preferred energy source, glucose, in the cell. This control is regulated by an allosteric regulatory protein, catabolite activator protein (CAP). When glucose levels in the cell are low, the organic molecule cyclic AMP is in high concentration.
Cyclic AMP activates CAP by binding to the allosteric sites, causing CAP to attach to the Lac operon promoter. Unlike the repressor proteins, binding of CAP to the Lac operon stimulates gene expression. When the cell glucose levels increase, the cyclic AMP levels in the cell decrease, and the activator protein will disassociate from the promoter. Transcription will return to low levels, or will turn off if the repressor protein reattaches.
Trp Operon
The Trp operon is responsible for synthesis of the amino acid trytophan when it is not available in the environment. The Trp operon is made up of a promoter with an operator, and five genes that encode enzymes for tryptophan synthesis. The Trp operon is regulated by the regulatory gene trpR, a gene that is located at a distance from the Trp operon.
The Trp operon is an example of a repressible operon; it is on unless turned off by a repressor protein. The repressor protein is synthesized by trpR. While the repressor protein is always present in the cell, it is synthesized in an inactive form. When a corepressor is present, in this case tryptophan, it binds to the repressor protein in an allosteric site.
This changes the conformation of the protein such that it can bind to the operator and block transcription by preventing the binding of RNA polymerase to the promoter. In this way the cell saves energy by not producing tryptophan when it is already present.
FAQ’s
An operon is a genetic regulatory system found in prokaryotic cells that coordinates the expression of multiple genes by controlling their transcription as a single unit.
An operon consists of a promoter region where RNA polymerase binds, an operator region where a regulatory protein binds to control transcription, and one or more structural genes that encode proteins with related functions. When the regulatory protein is active, it binds to the operator region and prevents RNA polymerase from binding to the promoter, blocking transcription of the structural genes. When the regulatory protein is inactive, it dissociates from the operator region, allowing RNA polymerase to bind to the promoter and initiate transcription of the structural genes.
There are two main types of operons: inducible operons and repressible operons. Inducible operons are normally off but can be turned on by a specific signal or substrate, while repressible operons are normally on but can be turned off by a specific signal or substrate. The lac operon, which controls the metabolism of lactose in E. coli, is an example of an inducible operon, while the trp operon, which controls the synthesis of tryptophan in E. coli, is an example of a repressible operon.
While operons are primarily found in prokaryotic cells, there are some examples of operon-like gene clusters in eukaryotic cells. For example, the Hox genes, which control the development of body segments in animals, are arranged in clusters on chromosomes and are co-regulated by enhancer elements that function similarly to operators. However, the regulation of gene expression in eukaryotic cells is generally more complex and involves a variety of additional mechanisms, including epigenetic modifications and the involvement of multiple regulatory proteins.